1 Fix_POSCAR(以atom_constrain.py为例)
1.1 atom_constrain.py 使用介绍
在对slab 结构进行计算时,有的时候为了减少计算量,可以考虑把slab 结构中某些层进行固定,目前固定的方法有很多,比如用MS进行手动固定,vaspkit-402 功能或许楠的POSCARtoolkit.py脚本等都可以实现不同方式的原子层固定,但目前vaspkit-402功能会把POSCAR坐标自动转换为笛卡尔坐标,POSCARtoolkit.py 功能虽然比较齐全,但操作比较复杂。因此,本脚本克服了上述问题,可以实现原本的POSCAR已经被固定或没被固定的情况,且输出文件不会对原有的POSCAR 进行转换。
脚本主要功能: 实现两种方式对POSCAR 进行选择性固定:
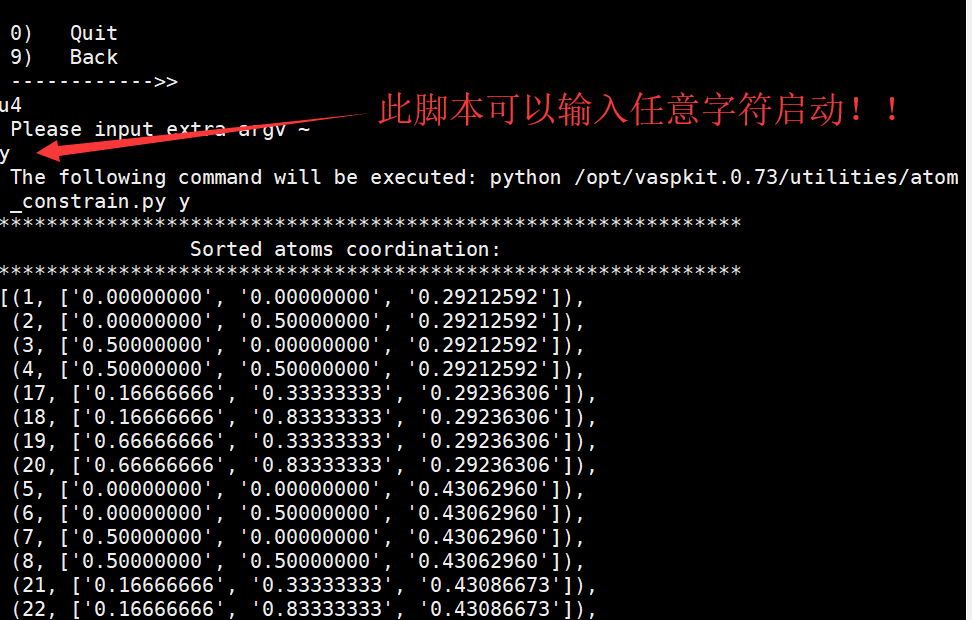
- 方式1:选择固定的总层数,适用于不对称表面的slab 结构,如图:
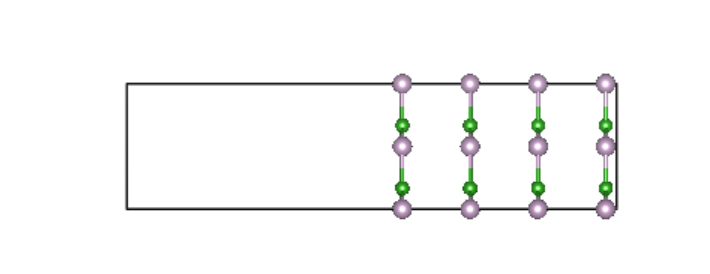
方式2:选择固定任意层数,适用于对称型表面
slab结构,如图:
下面详细介绍两种方式的使用流程。
方式1:
把atom_constrian.py 脚本拷贝到POSCAR 所在目录 然后python atom_constrain.py
屏幕将输出如下提示:
xxxxxxxxxx201**************************************************************2 Sorted atoms coordination: 3**************************************************************4[(2, ['2.315225840', '0.000000000', '0.000000000']),5 (3, ['0.000000000', '2.315225840', '0.000000000']),6 (7, ['3.472838759', '1.157612920', '1.157612920']),7 (8, ['1.157612920', '3.472838759', '1.157612920']),8 (1, ['0.000000000', '0.000000000', '2.315225840']),9 (4, ['2.315225840', '2.315225840', '2.315225840']),10 (5, ['1.157612920', '1.157612920', '3.472838759']),11 (6, ['3.472838759', '3.472838759', '3.472838759'])]12**************************************************************13 Please input a threshold value according to layer distance,14 the atoms coordination has been listed above.e.g:for direct a15 smaller value as 0.001,for cartesian a bigger value as 0.5 needed16 17**************************************************************1819Input:20 1脚本已经把POSCAR按照 z 方向坐标进行按层排序,并把所有原子序号及坐标打印到屏幕,由于原本的POSCAR 是笛卡尔坐标,所以z坐标单位是真实长度,且此时其层间距数值较大,如上面的结果所示,倒数第二层与最后一层间距数值差异达到1.1A,这个差值可以作为Input: 后面层间距的阈值判断的依据,输入的阈值在(0,1.1)范围内时,比如0.3或0.9、或1.1 等,脚本会自动把POSCAR 中两层间的距离大于输入的阈值的原子算作不同的层,比如此例中将会计算出4层,如果输入的阈值为1.2 或更大,由于POSCAR没有任何两层间的距离比1.2大,因此整个POSCAR 将会被脚本认为只有一层。
本例输入阈值为1,其输出结果如下,提示有4层被找到以及让用户输入方式1 或方式2 ,在这里输入a(选择方式1):`
xxxxxxxxxx1``` bash2**************************************************************3 There are total 4 layers are found!4**************************************************************5**************************************************************6 There are two methods to fix atoms in POSCAR:78The first one :input a total layer number,e.g. 5 means you will fix 5 layers from bottom to top9The second one:input a layer order number,e.g. 3 4 measn you10 will fix the 3th and 4th layer ,used in case where you want to fix11 center part of the POSCAR 12**************************************************************13**************************************************************1415 Please input a for method 1 and b for method 2:16a17**************************************************************18**************************************************************
如上面的输出结果所示,当选择方式1 后,提示输入固定的层数,此处输入3,并输入固定的类型为:F F F
xxxxxxxxxx11Please input the total number of layers to be fixed :32**************************************************************3**************************************************************45Please input which type you will fix,e.g. F F T,F F F,T F F etc.:6F F F7**************************************************************
最后脚本把被固定的原子的序数打印出来,并把被固定的结果输出到 POSCAR_NEW 里面。如下所示:
xxxxxxxxxx91**************************************************************2 Atoms number below have been fixed !34[2, 3, 7, 8, 1, 4]5 New POSCAR has been written in POSCAR_NEW !6**************************************************************7**************************************************************8 You may contact 316187631@QQ.com if any questions9**************************************************************注意:如果 输入a 或 F F F 提示有误,请输入'a' 和' F F F ' 替代!
方式2
使用方法和方式1基本相同,重新运行脚本,此次我故意选了一个分数坐标的 POSCAR
xxxxxxxxxx1171Zn4 O421.034.6304516792 0.0000000000 0.000000000040.0000000000 4.6304516792 0.000000000050.0000000000 0.0000000000 4.63045167926 Zn O7 4 48Selective Dynamics9Cartesian100.000000000 0.000000000 2.315225840 F F F112.315225840 0.000000000 0.000000000 F F F120.000000000 2.315225840 0.000000000 F F F132.315225840 2.315225840 2.315225840 F F F141.157612920 1.157612920 3.472838759 T T T153.472838759 3.472838759 3.472838759 T T T163.472838759 1.157612920 1.157612920 F F F171.157612920 3.472838759 1.157612920 F F F
x
1**************************************************************2 Sorted atoms coordination: 3**************************************************************4[(1, ['0.00000000', '0.00000000', '0.25641708']),5 (2, ['0.00000000', '0.50000000', '0.25641708']),6 (3, ['0.50000000', '0.00000000', '0.25641708']),7 (4, ['0.50000000', '0.50000000', '0.25641708']),8 (21, ['0.16666666', '0.33333333', '0.25682279']),9 (22, ['0.16666666', '0.83333333', '0.25682279']),10 (23, ['0.66666666', '0.33333333', '0.25682279']),11 (24, ['0.66666666', '0.83333333', '0.25682279']),12 (5, ['0.00000000', '0.00000000', '0.37810712']),13 (6, ['0.00000000', '0.50000000', '0.37810712']),14 (7, ['0.50000000', '0.00000000', '0.37810712']),15 (8, ['0.50000000', '0.50000000', '0.37810712']),16 (25, ['0.16666666', '0.33333333', '0.37851282']),17 (26, ['0.16666666', '0.83333333', '0.37851282']),18 (27, ['0.66666666', '0.33333333', '0.37851282']),19 (28, ['0.66666666', '0.83333333', '0.37851282']),20 (9, ['0.00000000', '0.00000000', '0.49979715']),21 (10, ['0.00000000', '0.50000000', '0.49979715']),22 (11, ['0.50000000', '0.00000000', '0.49979715']),23 (12, ['0.50000000', '0.50000000', '0.49979715']),24 (29, ['0.16666666', '0.33333333', '0.50020285']),25 (30, ['0.16666666', '0.83333333', '0.50020285']),26 (31, ['0.66666666', '0.33333333', '0.50020285']),27 (32, ['0.66666666', '0.83333333', '0.50020285']),28 (13, ['0.00000000', '0.00000000', '0.62148718']),29 (14, ['0.00000000', '0.50000000', '0.62148718']),30 (15, ['0.50000000', '0.00000000', '0.62148718']),31 (16, ['0.50000000', '0.50000000', '0.62148718']),32 (33, ['0.16666666', '0.33333333', '0.62189288']),33 (34, ['0.16666666', '0.83333333', '0.62189288']),34 (35, ['0.66666666', '0.33333333', '0.62189288']),35 (36, ['0.66666666', '0.83333333', '0.62189288']),36 (17, ['0.00000000', '0.00000000', '0.74317721']),37 (18, ['0.00000000', '0.50000000', '0.74317721']),38 (19, ['0.50000000', '0.00000000', '0.74317721']),39 (20, ['0.50000000', '0.50000000', '0.74317721']),40 (37, ['0.16666666', '0.33333333', '0.74358292']),41 (38, ['0.16666666', '0.83333333', '0.74358292']),42 (39, ['0.66666666', '0.33333333', '0.74358292']),43 (40, ['0.66666666', '0.83333333', '0.74358292'])]44**************************************************************45 Please input a threshold value according to layer distance,46 the atoms coordination has been listed above.e.g:for direct a47 smaller value as 0.001,for cartesian a bigger value as 0.5 needed48 49**************************************************************50Input:510.03注意:由于此时的Z方向数值差异较小,为0.12左右,因此 输入阈值在(0,0.12)之间均可。
接下来按提示输入:
x
123**************************************************************4 There are total 5 layers are found!5**************************************************************6**************************************************************7 There are two methods to fix atoms in POSCAR:89The first one :input a total layer number,e.g. 5 means you will fix 5 layers from bottom to top10The second one:input a layer order number,e.g. 3 4 measn you11 will fix the 3th and 4th layer ,used in case where you want to fix12 center part of the POSCAR 13**************************************************************14**************************************************************1516 Please input a for method 1 and b for method 2:17b18**************************************************************19**************************************************************2021Please input the number of layer order to be fixed :4 522**************************************************************23**************************************************************2425Please input which type you will fix,e.g. F F T,F F F,T F F etc.:26F F T27**************************************************************28**************************************************************29 Atoms number below have been fixed !3031[13, 14, 15, 16, 33, 34, 35, 36, 17, 18, 19, 20, 37, 38, 39, 40]32 New POSCAR has been written in POSCAR_NEW !33**************************************************************34**************************************************************35 You may contact 316187631@QQ.com if any questions36**************************************************************我选择了方式2,输入b ,层数输入4 5,并输入F F T 固定类型,表示 我将要把从底部往上的第一层
和第二层进行相应的固定。输出结果同样把被固定的原子数打印出来。其POSCAR_NEW显示如下:
491Super Cell Generated by VASPKIT20.837.99480009 0.00000000 0.000000004-3.99740005 6.92369998 0.0000000050.00000000 0.00000000 30.815999986 P B7 20 208Selective Dynamics9Direct100.00000000 0.00000000 0.25641708 T T T110.00000000 0.50000000 0.25641708 T T T120.50000000 0.00000000 0.25641708 T T T130.50000000 0.50000000 0.25641708 T T T140.00000000 0.00000000 0.37810712 T T T150.00000000 0.50000000 0.37810712 T T T160.50000000 0.00000000 0.37810712 T T T170.50000000 0.50000000 0.37810712 T T T180.00000000 0.00000000 0.49979715 T T T190.00000000 0.50000000 0.49979715 T T T200.50000000 0.00000000 0.49979715 T T T210.50000000 0.50000000 0.49979715 T T T220.00000000 0.00000000 0.62148718 F F T230.00000000 0.50000000 0.62148718 F F T240.50000000 0.00000000 0.62148718 F F T250.50000000 0.50000000 0.62148718 F F T260.00000000 0.00000000 0.74317721 F F T270.00000000 0.50000000 0.74317721 F F T280.50000000 0.00000000 0.74317721 F F T290.50000000 0.50000000 0.74317721 F F T300.16666666 0.33333333 0.25682279 T T T310.16666666 0.83333333 0.25682279 T T T320.66666666 0.33333333 0.25682279 T T T330.66666666 0.83333333 0.25682279 T T T340.16666666 0.33333333 0.37851282 T T T350.16666666 0.83333333 0.37851282 T T T360.66666666 0.33333333 0.37851282 T T T370.66666666 0.83333333 0.37851282 T T T380.16666666 0.33333333 0.50020285 T T T390.16666666 0.83333333 0.50020285 T T T400.66666666 0.33333333 0.50020285 T T T410.66666666 0.83333333 0.50020285 T T T420.16666666 0.33333333 0.62189288 F F T430.16666666 0.83333333 0.62189288 F F T440.66666666 0.33333333 0.62189288 F F T450.66666666 0.83333333 0.62189288 F F T460.16666666 0.33333333 0.74358292 F F T470.16666666 0.83333333 0.74358292 F F T480.66666666 0.33333333 0.74358292 F F T490.66666666 0.83333333 0.74358292 F F T
1.2 把atom_constrain.py 加入vaspkit 统一管理
Vaspkit 目前功能越来越完善,如果用户自己有些小的脚本可以补充vaspkit的功能,那么,可以把自己的小脚本“集成到”vaspkit里面,用vaspkit统一管理自己的脚本,现在以atom_constrain.py为例讲解如何把该脚本“放到”vaspkit 里面,利用vaspkit 惯有的工作模式进行使用:
第一步:进入vaspkit安装目录下的utilities 目录,我安装vaspkit的路径如下 :
xxxxxxxxxx11/public/home/vaspkit.0.73/utilities第二步:把atom_constrain.py 拷贝到此
第三步:vim ~/.vaspkit (或vi~/.vaspkit)
按图下修改:注意图中箭头处
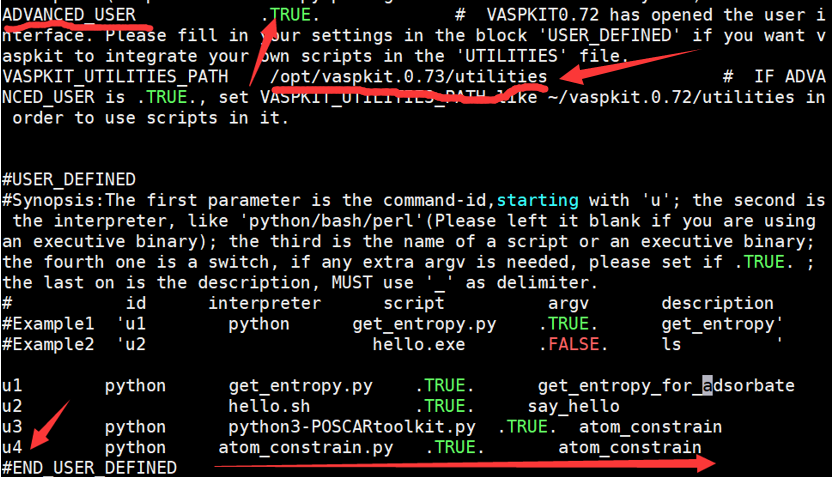
输入“:wq” ,保存并退出
第四步,进入含有POSCAR 的目录体验一把:
输入vaspkit--->>74得到如下图示:
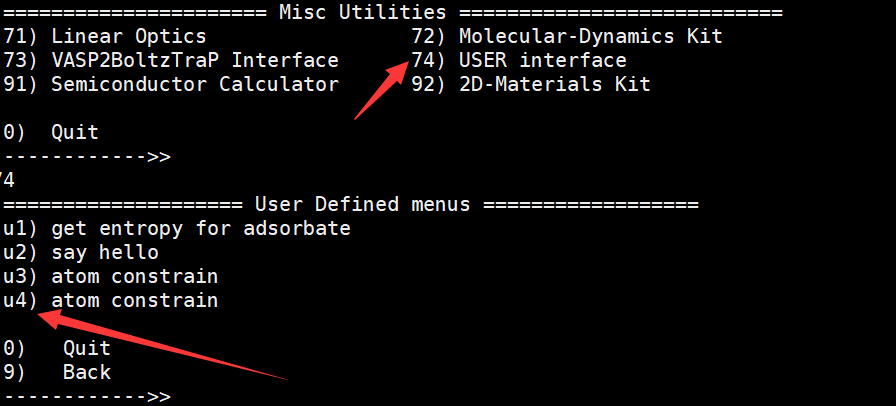
接着输入u4 ,再输入任何字符回车后进入脚本工作页面: